1.NBS gene:
============
Nijmegen breakage syndrome
From Wikipedia, the free encyclopedia
| Nijmegen breakage syndrome | |
|---|---|
| Classification and external resources | |
| OMIM | 251260 |
| DiseasesDB | 32395 |
| eMedicine | derm/725 |
| MeSH | D049932 |
NBS1 codes for a protein that has two major functions: (1) to stop the cell cycle in the S phase, when there are errors in the cell DNA (2) to interact with FANCD2 that can activate the BRCA1/BRCA2 pathway of DNA repair. This explains clearly that mutations in the NBS1 gene lead to higher levels of cancer (see Fanconi anemia, Cockayne syndrome...)
The name derives from the Dutch city Nijmegen where the condition was first described.[2]
Most people with NBS have West Slavic origins. The largest number of them live in Poland.
Mrs Seemanova MD after whom the name of the syndrome was given, currently works at Motol Hospital, Prague, Czech Republic, as a Professor of medical genetics.===============================================
2. BLM gene
Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.[1]
The Bloom syndrome gene product is related to the RecQ subset of DExH box-containing DNA helicases and has both DNA-stimulated ATPase and ATP-dependent DNA helicase activities. Mutations causing Bloom syndrome delete or alter helicase motifs and may disable the 3' → 5' helicase activity. The normal protein may act to suppress inappropriate homologous recombination.[2]
Interactions
Bloom syndrome protein has been shown to interact with CHEK1,[3] Replication protein A1,[4][5][6] Werner syndrome ATP-dependent helicase,[7] RAD51L3,[8] Ataxia telangiectasia mutated,[9][10] RAD51,[11] XRCC2,[8] Flap structure-specific endonuclease 1,[12] H2AFX,[3] TP53BP1,[3] FANCM,[13] P53,[3][14][15][16] TOP3A,[4][17][18][19] MLH1[9][18][20][21] and CHAF1A.[22]TO UNDERSTAND THAT THE BLM GENE IS A DNA REPAIR GENE, YOU MUST REMEMBER WHAT A 'HELICASE' IS:
Helicase
From Wikipedia, the free encyclopedia
| DNA helicase | |||
|---|---|---|---|
| Identifiers | |||
| EC number | 3.6.4.12 | ||
| Databases | |||
| IntEnz | IntEnz view | ||
| BRENDA | BRENDA entry | ||
| ExPASy | NiceZyme view | ||
| KEGG | KEGG entry | ||
| MetaCyc | metabolic pathway | ||
| PRIAM | profile | ||
| PDB structures | RCSB PDB PDBe PDBsum | ||
|
|||
| RNA helicase | |||
|---|---|---|---|
| Identifiers | |||
| EC number | 3.6.4.13 | ||
| Databases | |||
| IntEnz | IntEnz view | ||
| BRENDA | BRENDA entry | ||
| ExPASy | NiceZyme view | ||
| KEGG | KEGG entry | ||
| MetaCyc | metabolic pathway | ||
| PRIAM | profile | ||
| PDB structures | RCSB PDB PDBe PDBsum | ||
|
|||
KEEFE ET AL" Bloom syndrome occurs most frequently in the Ashkenazi Jewish population with patients almost exclusively homozygous for a frameshift mutation resulting from a 6 bp deletion/7 bp insertion at nucleotide 2,281 (BLMAsh). This mutation causes premature termination of the encoded gene product producing a truncated protein of 739 amino acids while the full length protein contains 1417 amino acids.
The mutated gene in Bloom syndrome, BLM, was localized to chromosome 15q26.1 and encodes a member of the RecQ family of DNA helicases. This family also contains several other genes that are associated with disease phenotypes including the Werner Syndrome protein (WRN) and the defective protein in Rothmund-Thomson syndrome (RecQL4). Both of these diseases also feature an increased incidence of cancer. BLM, along with the rest of the members of this family, exhibits 3'-5' helicase activity and plays a role in DNA repair and recombination. BLM functions during replication stress and is required for the recruitment of several other important repair proteins including NBS1, BRCA1, Rad51 and MLH1. In addition, the BLM helicase is involved in recombinational repair events as evidenced by its ability to promote branch migrations of Holliday junctions at stalled replication forks. BLM may also play a role in apoptosis since it directly interacts with p53 and helps regulate its transcriptional activity."
=========================================================================
3.ATM gene
Ataxia telangiectasia mutated
From Wikipedia, the free encyclopedia
| Ataxia telangiectasia mutated | |||||
|---|---|---|---|---|---|
| Identifiers | |||||
| Symbols | ATM; AT1; ATA; ATC; ATD; ATDC; ATE; TEL1; TELO1 | ||||
| External IDs | OMIM: 607585 MGI: 107202 HomoloGene: 30952 ChEMBL: 3797 GeneCards: ATM Gene | ||||
| EC number | 2.7.11.1 | ||||
|
|||||
| Orthologs | |||||
| Species | Human | Mouse | |||
| Entrez | 472 | 11920 | |||
| Ensembl | ENSG00000149311 | ENSMUSG00000034218 | |||
| UniProt | Q13315 | Q62388 | |||
| RefSeq (mRNA) | NM_000051 | NM_007499 | |||
| RefSeq (protein) | NP_000042 | NP_031525 | |||
| Location (UCSC) | Chr 11: 108.09 – 108.24 Mb |
Chr 9: 53.44 – 53.54 Mb |
|||
| PubMed search | [1] | [2] | |||
The protein is named for the disorder Ataxia telangiectasia caused by mutations of ATM.[1]
GOLDGAR ET AL suggested:
"The risk estimates from this study suggest that women carrying the pathogenic variant, ATM c.7271T > G, or truncating mutations demonstrate a significantly increased risk of breast cancer with a penetrance that appears similar to that conferred by germline mutations in BRCA2."
=============================================================4. MRE 11 gene
MRE11A
From Wikipedia, the free encyclopedia
| MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||
| Identifiers | |||||||||||
| Symbols | MRE11A; ATLD; HNGS1; MRE11; MRE11B | ||||||||||
| External IDs | OMIM: 600814 MGI: 1100512 HomoloGene: 4083 GeneCards: MRE11A Gene | ||||||||||
|
|||||||||||
| RNA expression pattern | |||||||||||
 |
|||||||||||
| More reference expression data | |||||||||||
| Orthologs | |||||||||||
| Species | Human | Mouse | |||||||||
| Entrez | 4361 | 17535 | |||||||||
| Ensembl | ENSG00000020922 | ENSMUSG00000031928 | |||||||||
| UniProt | P49959 | Q61216 | |||||||||
| RefSeq (mRNA) | NM_005590 | NM_018736 | |||||||||
| RefSeq (protein) | NP_005581 | NP_061206 | |||||||||
| Location (UCSC) | Chr 11: 94.15 – 94.23 Mb |
Chr 9: 14.78 – 14.84 Mb |
|||||||||
| PubMed search | [1] | [2] | |||||||||
This gene encodes a nuclear protein involved in homologous recombination, telomere length maintenance, and DNA double-strand break repair. By itself, the protein has 3' to 5' exonuclease activity and endonuclease activity. The protein forms a complex with the RAD50 homolog; this complex is required for nonhomologous joining of DNA ends and possesses increased single-stranded DNA endonuclease and 3' to 5' exonuclease activities. In conjunction with a DNA ligase, this protein promotes the joining of noncomplementary ends in vitro using short homologies near the ends of the DNA fragments. This gene has a pseudogene on chromosome 3. Alternative splicing of this gene results in two transcript variants encoding different isoforms.[2]
Interactions
MRE11A has been shown to interact with Ku70,[3] Ataxia telangiectasia mutated,[4][5] MDC1,[6] Rad50,[3][5][7][8][9] Nibrin,[5][9][10][11][12] TERF2[13] and BRCA1.[5][7][14][15]FUKUDA ET AL
"MRE11, RAD50, and XRS2 have been identified in yeast as components of the HR and NHEJ pathways (4) . A physical complex with these proteins has been identified. In vertebrates, MRE11 and RAD50 form a complex with NBS1, whose mutation causes NBS (5 , 6) . The clinical features of NBS overlap with those of AT. They are characterized by chromosome instability, increased hypersensitivity to ionizing radiation, immunodeficiency, and predisposition to cancer. AT is caused by mutations in the ATM gene, which encodes a protein kinase homologous with phosphatidylinositol-3 kinase (7) . ATM is a key regulator of the cellular response to DSBs. NBS1 is phosphorylated in an ATM-dependent manner after ionizing radiation, suggesting a link between ATM and NBS1 in a common signaling pathway (8) . MRE11 phosphorylation upon DNA damage is dependent on NBS1 (9) . Therefore, it is highly likely that MRE11 participates in the same pathway in response to DNA damage. Consistent with this functional interaction, hypomorphic mutations in the MRE11 gene cause ataxia-telangiectasia-like disorder, the phenotypes of which are indistinguishable from those of AT (10) ."
ATM mutations play a causal role in AT and have been demonstrated in lymphoid malignancies
======================================================================
5. RAD51
From Wikipedia, the free encyclopedia
| RAD51 homolog (S. cerevisiae) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
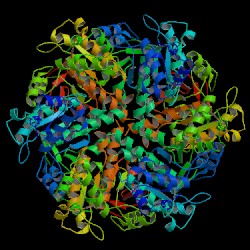 A filament of Rad51 based on PDB 1SZP.[1] |
|||||||||||
|
|||||||||||
| Identifiers | |||||||||||
| Symbols | RAD51; BRCC5; HRAD51; HsRad51; HsT16930; MRMV2; RAD51A; RECA | ||||||||||
| External IDs | OMIM: 179617 MGI: 97890 HomoloGene: 2155 GeneCards: RAD51 Gene | ||||||||||
|
|||||||||||
| RNA expression pattern | |||||||||||
 |
|||||||||||
 |
|||||||||||
| More reference expression data | |||||||||||
| Orthologs | |||||||||||
| Species | Human | Mouse | |||||||||
| Entrez | 5888 | 19361 | |||||||||
| Ensembl | ENSG00000051180 | ENSMUSG00000027323 | |||||||||
| UniProt | Q06609 | Q08297 | |||||||||
| RefSeq (mRNA) | NM_001164269 | NM_011234 | |||||||||
| RefSeq (protein) | NP_001157741 | NP_035364 | |||||||||
| Location (UCSC) | Chr 15: 40.99 – 41.02 Mb |
Chr 2: 119.11 – 119.15 Mb |
|||||||||
| PubMed search | [1] | [2] | |||||||||
BRCA genes
This protein can interact with the ssDNA-binding protein RPA, BRCA2, PALB2[3] and RAD52.
The structural basis for Rad51 filament formation and its functional mechanism still remain poorly understood. However, recent studies using fluorescent labeled Rad51[4] has indicated that Rad51 fragments elongate via multiple nucleation events followed by growth, with the total fragment terminating when it reaches about 2 μm in length. Disassociation of Rad51 from dsDNA, however, is slow and incomplete, suggesting that there is a separate mechanism that accomplishes this."
"The RAD51 gene family, genetic instability and cancer.
Source
Medical Research Council, Radiation and Genome Stability Unit, Harwell, Oxfordshire OX11 0RD, UK. j.thacker@har.mrc.ac.ukAbstract
Inefficient repair or mis-repair of DNA damage can cause genetic instability, and defects in some DNA repair genes are associated with rare human cancer-prone disorders. In the last few years, homologous recombination has been found to be a key pathway in human cells for the repair of severe DNA damage such as double-strand breaks. The RAD51 family of genes, including RAD51 and the five RAD51-like genes (XRCC2, XRCC3, RAD51L1, RAD51L2, RAD51L3) are known to have crucial non-redundant roles in this pathway."---------------------------------------------------------------------THE BRCA ITSELF TO WORK NEED A BUNCH OF VARIOUS COFACTORS CALLED FANCB-Related Fanconi Anemia, FANCC-Related Fanconi Anemia, FANCD2-Related Fanconi Anemia, FANCE-Related Fanconi Anemia, FANCF-Related Fanconi Anemia, FANCG-Related Fanconi Anemia, FANCI-Related Fanconi Anemia, FANCL-Related Fanconi Anemia, FANCM-Related Fanconi Anemia, PALB2-Related Fanconi Anemia, RAD51C-Related Fanconi Anemia, SLX4-Related Fanconi Anemia (ALTER ET AL!)
========================================================================
IN OTHER NEWS...